Mukerji, Oliver Co-Author Study in PNAS on Basic Cell Function
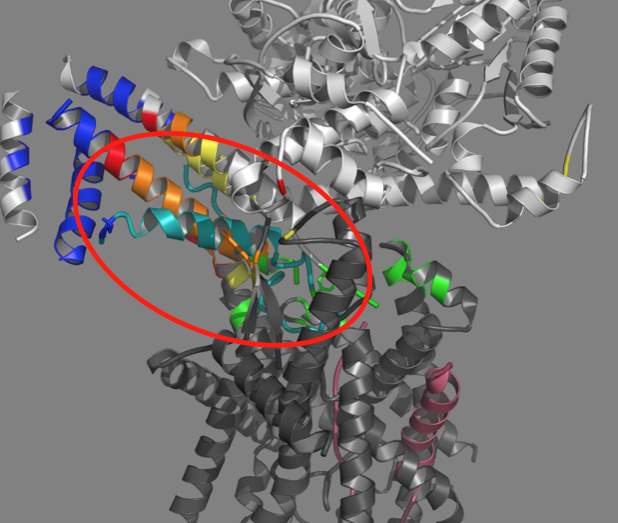
All cells — bacterial or human — secrete up to 10 or 20 percent of the proteins that they make. Human secreted proteins, for example, include components of serum, hormones, growth factors that promote cell development during embryogenesis and tissue remodeling, and proteins that provide the basis for immune cell signaling during infection or when fighting cancer.
The secretion process, however, isn’t an easy feat for cells, as they need to move the proteins across a membrane through a channel. Transport requires the formation of a hairpin, formed by an initiator protein.
In a recent study, Don Oliver, the Daniel Ayres Professor of Biology, professor of molecular biology and biochemistry, and Ishita Mukerji, the Fisk Professor of Natural Science, professor of molecular biology and biochemistry, explain the importance of where and why hairpins form and how they help proteins move across the cell.
The study, titled “Alignment of the protein substrate hairpin along the SecA two-helix finger primes protein transport in Escherichia coli,” brings together key areas of membrane biochemistry, structural biology and molecular biophysics, and has innovative applications of molecular genetics and fluorescence spectroscopy. It was published in the Aug. 7 issue of Proceedings of the National Academy of Sciences (PNAS).
Qi Zhang PhD ’16; Zhongmou Sun ’16; and molecular biology and biochemistry graduate students Sudipta Lahiri and Tithi Banerjee also contributed to the research.
SecA and SecYEG are protein complexes that are involved in transporting proteins across membranes. SecA brings the protein to the channel (SecYEG). Prior to this study, it was thought that the hairpin only formed within the channel.
“Fundamental questions remain regarding when and how this hairpin structure forms,” Mukerji explained. “What was really exciting in this study is that we saw evidence for the hairpin forming prior to insertion into the channel.”
The hairpin observed in the study contains the molecular ‘zipcode’ or signal sequence that contains the information about where the protein needs to go in the cell. Their study provides some insight as to how the zipcode is read both by SecA and SecYEG.
“Given that bacteria are becoming resistant to our current range of antibiotics, it is always important to identify new potential targets that are unique to bacteria,” Mukerji explained. “The SecA-SecYEG system can be a target for antibiotics and thus, understanding the system better can lead to a better and more effective design for therapeutics.”

